Original Article
Author Details :
Volume : 5, Issue : 1, Year : 2019
Article Page : 18-22
https://doi.org/10.18231/2581-4761.2019.0005
Abstract
Introduction: In 1899 Thiercelin first described ‘Enterococcus’. They are primarily member of gastrointestinal flora of human and animals. Enterococcus was grouped under streptococcus as group D streptococci but now it has been separated as individual genus. Clinically the most important species associated with human Infections are E. faecalis and E. faecium. They are frequently resistant to various group of antimicrobials. UTI is the most common infection among all Enterococcal infections and implicated in approximately 10 percent of all UTIs.
Materials and Methods: The samples were collected from all hospitalized and OPD patients of MBS Hospital, JK Lone Hospital & NMC Hospital. Government Medical College, Kota, Rajasthan. A total of 360 isolates were collected during the period of 2 years from April 2016 to April 2018 and processed at Microbiology Laboratory, Department of Microbiology, Government Medical College, Kota, Rajasthan.
Results: The present study shows a high incidence of enterococcal UTI among females (225) compare to males (135) with maximum Isolation among population between age group of 31- 40 (111) followed by 41-50 (59) and 21-30 (48). In present study the Ampicillin, Piperacillin, Ciprofloxacin, Norfloxacin, Nitrofurantoin, Gentamicin, Vancomycin and Linezolid antibiotic discs was tested and Linezolid was found to be most effective among all followed by Vancomycin, Nitrofurantoin, Ampicillin, Piperacillin, Ciprofloxacin, Norfloxacin. High level Gentamicin resistance (HLAR) was 60.27% in present study and Vancomycin resistance was 14.16 %.
Conclusion: Among all species of enterococci E. faecium were most resistant species among all. Ampicillin & Piperacillin resistance was considerably higher in present study in comparison to other studies. Resistance to Nitrofurantoin was also found highest among E. faecium 77.77% in present study and other compared studies.[1] By observing the various parameters of present study it can be concluded that enterococci which was thought to be a commensal organism is now emerging as a potential pathogen, particularly among hospitalised patients. In our institution Enterococcus isolates was more resistant to fluoroquinolones, aminoglycosides and ?-lactams agents. Early detection of Enterococcal species and resistance to Aminoglycoside and Vancomycin can be helpful in limiting the morbidity in our hospital set up.
Keywords: Enterococcus, UTI, Antibiotic resistance.
In 1899 Thiercelin described a saprophytic coccus from intestinal origin of almost all animals and term it "Enterococcus".[2] Enterococci are primarily member of gastrointestinal flora of human and animals.[2],[3]Earlier the Enterococcus was grouped under streptococcus as group D streptococci but in past decades Enterococcus group has undergone various taxonomic changes and recognizes it as a separate genus, as it show no similar homology with streptococcus genus.[2],[4]The current criteria of including bacteria in genus Enterococcus is based completely on molecular basis like DNA-DNA recognition experiments, 16 S RNA gene sequencing, analysis of whole cell protein profile etc.[2],[5]Enterococcus genus has been phenotypically divided by Facklam & collaborators in 2002 into 5 groups based on acid formation in Mannitol & Sorbose broth as well as arginine hydrolysis.[2],[6]Clinically the most important species associated with human Infections are E. faecalis and E. faecium.[7] Of these, E. faecalis is the most pathogenic species but E. faecium is of growing importance as it is more frequently resistant to antimicrobials.[8] Antimicrobial resistance can be classified as either intrinsic or acquired. Intrinsic resistance is related to inherent or natural chromosomally encoded characteristics present in all or most of the Enterococci. Enterococcal intrinsic resistance involves two major groups of antimicrobial therapeutic drugs: the aminoglycosides and ?-lactams.[9] In addition to the intrinsic resistance traits, Enterococci have acquired different genetic determinants that confer resistance to several classes of antibiotics, including Chloramphenicol, Tetracycline, Macrolides, lincosamides, and Streptogramins, Aminoglycosides. Antibiotic resistance is a major factor involved in nosocomial infections. It allows the organisms of low pathogenicity like Enterococci, to endure in the environment in which antimicrobial agents are heavily used thereby providing a selective advantage. Enterococci have become the second most common agent recovered from nosocomial urinary tract infections (UTI). UTI is the most common infection among all Enterococcal infections; Enterococci have been implicated in approximately 10 percent of all UTIs.[10]
The samples were collected from all hospitalized and OPD patients of MBS hospital, JK Lone Hospital & NMC hospital. Government Medical College, Kota, Rajasthan. A total of 360 isolates were collected during the period of 2 years from April 2016 to April 2018 and processed at Microbiology Laboratory, Department of Microbiology, Government Medical College, Kota, Rajasthan. All the isolates were identified as enterococci by gram staining, Motility, Bile esculin hydrolysis, salt tolerance test and further up to species level by following standard identification scheme by G.I. Barrow.[11]
All the isolates identified as enterococci were tested for their antibiotic susceptibility using Kirby-Bauer disc diffusion method and interpreted according to CLSI guidelines.[12],[13]Antibiotic discs (Himedia) used for testing susceptibility of enterococcal isolates were Ampicillin 10 µg, Piperacillin 75 µg, Ciprofloxacin 5 µg, Norfloxacin 10 µg, Nitrofurantoin 300 µg, Vancomycin 30 µg, Linezolid 30 µg, Gentamicin high content 120 µg.
 |
Click here to view |
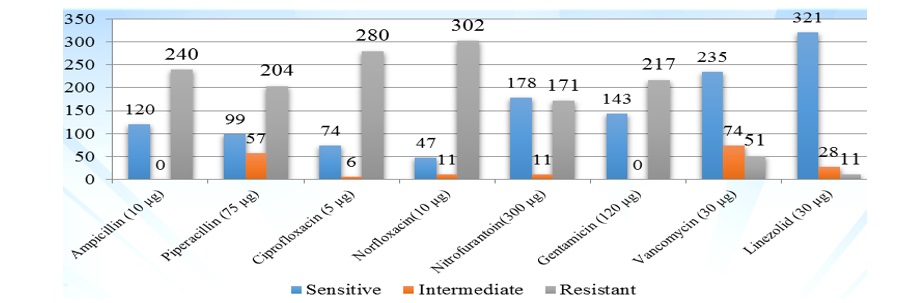
In present study the Ampicillin, Piperacillin, Ciprofloxacin, Norfloxacin, Nitrofurantoin, Gentamicin, Vancomycin and Linezolid antibiotic discs was tested by Kirby-Bauer disc diffusion method. Linezolid was most effective among all showed sensitivity for 321(89.16%), intermediate to 28(7.77%) and resistance for 11(3.05%) isolates, followed by vancomycin sensitive to 235(65.27%) intermediate to 74(20.55%) and resistance for 51(14.16 %) isolates, Nitrofurantoin sensitive to 178(49.44%) intermediate to 11(3.05%) and resistance for 171(47.5%) isolates, Ampicillin sensitive to 120(33.33%) and resistance for 240(66.66%) isolates, Piperacillin sensitive to 99(27.5%) intermediate to 57(15.83%) and resistance for 204(56.66%) isolates, Ciprofloxacin sensitive to 74(20.55%) intermediate to 6(1.66%) and resistance for 280(77.77%) isolates, Norfloxacin sensitive to 47(13.05%) intermediate to 11(3.05%) and resistance for 302(83.88%) isolates. High level Gentamicin was tested for HLAR detection, this was sensitive to 143(39.72%) and resistance for 217(60.27%) isolates (Fig. 2).
Results
In the present study samples were processed in department of Microbiology Govt. Medical College Kota. 360 isolates of enterococci were recovered in present study and distribution of isolates from samples collected from patients of both OPD and IPD was 256 and 104. A total 43 isolates was from catheterized patients among both OPD & IPD.
The present study shows a high incidence of enterococcal UTI among females (225) compare to males (135) so Male to Female ratio was 0.6. Isolation of enterococci was maximum among population between age group of 31- 40 (111) followed by 41-50 (59) and 21-30 (48) (Table-1) (Fig. 1).
Table 1: Distribution of enterococcus UTI in various age and sex
|
Age |
Male |
Female |
Total |
|
0-10 |
04 |
16 |
20 |
|
11-20 |
10 |
15 |
25 |
|
21-30 |
16 |
32 |
48 |
|
31-40 |
37 |
74 |
111 |
|
41-50 |
28 |
31 |
59 |
|
51-60 |
19 |
24 |
43 |
|
61-70 |
10 |
20 |
30 |
|
71and above |
11 |
13 |
24 |
|
Total |
135 |
225 |
360 |
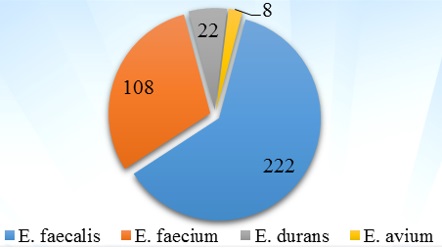
Most common species found in present study was Enterococcus faecalis 222 (61.66%) followed by E. faecium108 (30%), E. durans 22 (6.11%) and E. avium 8 (2.22%)(Fig. 1).
 |
Click here to view |
Fig. 2: Antibiotic sensitivity pattern among all enterococcus by Kirby -Bauer method
Antibiotics sensitivity pattern among different Enterococcus Species
Antibiotic susceptibility pattern among various enterococcus species has been shown in following Table-2. E. faecalis show highest sensitivity for Linezolid 195(87.83%) followed by Vancomycin 159(71.62%), Nitrofurantoin 136(61.26%), Ampicillin 80(36.03%), Piperacillin 73(32.88%), Ciprofloxacin 54(24.32%) and least for Norfloxacin (37(16.66%).
E. faecium show highest sensitivity for Linezolid 98 (90.74%) followed by Vancomycin 51(47.22%), Ampicillin 25(23.14%), Piperacillin 24(22.22%), Nitrofurantoin 22(20.37%), Ciprofloxacin and Norfloxacin show equal sensitivity 7(6.48%).
E. durans show highest sensitivity for Linezolid 22(100%) followed by Vancomycin 20(90.9%), Nitrofurantoin 15(68.18%), Ampicillin and Piperacillin show equal sensitivity 11(50%), Norfloxacin 7(31.81%), and least for Ciprofloxacin 5(22.72%).
E. avium showed highest sensitivity for Linezolid 6(75%) followed by vancomycin 5(62.5%), ampicillin, Piperacillin and Nitrofurantoin show same result 4(50%) and least for ciprofloxacin and Norfloxacin 2(25%).
High level Gentamicin was used to detect high level resistance for aminoglycosides among all enterococcus species E. faecium isolates show maximum resistance for aminoglycoside 92(85.18%), followed by E. faecalis 120(54.05%), E. avium 2(25%) and E. durans 3(13.63%) where the N value was 108, 222, 8, and 22 respectively.
Table 2: Sensitivity and resistance pattern to various antibiotics in different Enterococcus Species
|
Enterococcus spp. |
|
Amp* (%) |
Pip* (%) |
Cip* (%) |
Nor* (%) |
Nit* (%) |
Van* (%) |
Lz* (%) |
HLG* (%) |
|
E. faecalis N=222 |
S |
80 (36.03) |
73 (32.88) |
54 (24.32) |
37 (16.66) |
136 (61.26) |
159 (71.62) |
195 (87.83) |
102 (45.94) |
|
I |
0 (0) |
33 (14.86) |
7 (3.15) |
1 (0.45) |
8 (3.6) |
44 (19.81) |
23 (10.36) |
0 (0) |
|
|
R |
142 (63.96) |
116 (52.25) |
161 (72.52) |
184 (82.88) |
78 (35.13) |
19 (8.55) |
4 (1.8) |
120 (54.05) |
|
|
E. faeciumN=108 |
S |
25 (23.14) |
24 (22.22) |
7 (6.48) |
7 (6.48) |
22 (20.37) |
51 (47.22) |
98 (90.74) |
16 (14.81) |
|
I |
0 (0) |
7 (6.48) |
3 (2.77) |
0 (0) |
2 (1.85) |
27 (25) |
4 (3.7) |
0 (0) |
|
|
R |
83 (76.85) |
77 (71.29) |
98 (90.74) |
101 (93.51) |
84 (77.77) |
30 (27.77) |
6 (5.55) |
92 (85.18) |
|
|
E. durans N=22 |
S |
11 (50) |
11 (50) |
5 (22.72) |
7 (31.81) |
15 (68.18) |
20 (90.9) |
22 (100) |
19 (86.36) |
|
I |
0 (0) |
2 (9.09) |
2 (9.09) |
1 (4.54) |
1 (4.54) |
2 (9.09) |
0 (0) |
0 (0) |
|
|
R |
11 (50) |
9 (40.9) |
15 (68.18) |
14 (63.63) |
6 (27.27) |
0 (0) |
0 (0) |
3 (13.63) |
|
|
E. avium N=8 |
S |
4 (50) |
4 (50) |
2 (25) |
2 (25) |
4 (50) |
5 (62.5) |
6 (75) |
6 (75) |
|
I |
0 (0) |
2 (25) |
0 (0) |
1 (12.5) |
1 (12.5) |
1 (12.5) |
1 (12.5) |
0 (0) |
|
|
R |
4 (50) |
2 (25) |
6 (75) |
5 (62.5) |
3 (37.5) |
2 (25) |
1 (12.5) |
2 (25) |
*Amp=Ampicillin, Pip=Piperacillin, Cip=Ciprofloxacin, Nor=Norfloxacin, Nit=Nitrofurantoin, Van=Vancomycin, Lz=Linezolid, HLG=High level Gentamicin.
Enterococcus species are continuously emerging as important pathogen specially in hospital environment and can cause different type of infection which are usually difficult to treat due to limited antibiotic options and higher incidence of drug resistance to various antibiotics. Enterococcus is a most important cause of urinary tract infection (UTI) caused by gram positive bacteria.
In the present study 360 urinary isolates of enterococci were studied during the period of 2 years from April 2016 to April 2018, among them more cases were from females (225) then males (135) which could be due to close proximity of anal orifice and urethra in females or due to poor hygiene.
E. faecalis: Among all groups of antibiotics E. faecalis show highest resistance for Fluoroquinolones in comprision to other study Norfloxacin resistance was higher in present study. Ampicillin resistance in present study is higher compared to other studies. Resistance to Nitrofurantoin is also higher among E. faecalis in present study compared to study done by varun goel et al.
Aminoglycoside resistance was detected by Gentamicin 120 µg disc, in present study E. faecalis show considerably high resistance to Gentamicin the result is similar to study of Sanal C. Fernandes et al.
Natural intrinsic tolerance to Aminoglycosides has been shown by enterococci. This property is due to two main factors, poor entry of antibiotic and inactivation of antibiotic by covalent modification of the hydroxyl or amino groups by naturally occurring enterococcal enzymes. In addition to this enterococci can modify the ribosomal target by the action of ribosomal RNA (rRNA) methyltransferase known as EfmM.[14] ,[15] Vancomycin resistance among E. faecalis in present study is comparatively higher than a study from south India done by Sanal C. Fernandes et al and lower from study of north India AIIMS done by varun goel et al
Linezolid resistance among E. faecalis in present study was 1.8% which is higher than other studies.
E. faecium: Among all species of enterococci E. faecium were most resistant species among all. They show highest resistance to Fluoroquinolone, followed by Aminoglycosides, Nitrofurantoin, Ampicillin, Piperacillin, Vancomycin and Linezolid.
Fluoroquinolone resistance among E. faecalis was highest then other species and comparatively it was lower than other studies of Yaeghob Sharifi et al, Saraswathy MP et al. and was higher than study of Varun goel et al.
E. faecium shares all resistance mechanism with E. faecalis in addition efflux pump mediated by NorA gene.[14],[16],[17]
Ampicillin resistance was considerably higher in present study as compare to most of studies except study done by Yaeghob Sharifi et al which showed higher resistance than present study, similarly Piperacillin resistance was also high 71.29% in present study. Resistance to Nitrofurantoin was found highest among E. faecium 77.77% in present study and other compared studies.[18]
Gentamicin resistance for E. faecium was 85.18% also highest compared to other species in present and other studies.[19] ,[1]Vancomycin resistance among E. faecium 27.77% which is higher than other studies.[18] ,[20]Linezolid resistance was highest among E. faecium then other species of enterococci in this study.
E. durans and E. avium was isolated in lesser amount and E. avium was more resistant to almost all groups of antibiotics studied. As the number of isolates of E. durans and E. avium was less so significant comparison can not be done.
Conclusion
Enterococci is a notorious pathogen as it has intrinsic resistance/tolerance to different group of antibiotics and is a main causative agent of gram positive UTI. By observing the various parameters of present study it can be concluded that enterococci which was thought to be a commensal organism is now emerging as a potential pathogen, particularly among hospitalised patients. E. faecalis and E. faecium found to be the most prevalent species which confer resistance to various groups of antibiotics. E. faecium found to be more resistant species then E. faecalis. In our institution Enterococcus isolates was more resistant to fluoroquinolones, aminoglycosides and ?-lactams agents like Ampicillin and Piperacillin. This may be due to selection pressure of these antibiotics in our set up. Early detection of Enterococcal species and resistance to Aminoglycoside and Vancomycin can be helpful in limiting the morbidity in our hospital set up.
Conflicts of Interest: None.
How to cite : Naruka H S, Chand A E, Meena H, Prevalence of various enterococcus species and their antibiotic resistance pattern among urinary isolates in tertiary care center in South Eastern Rajasthan. IP Int J Med Microbiol Trop Dis 2019;5(1):18-22
This is an Open Access (OA) journal, and articles are distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
Viewed: 3695
PDF Downloaded: 635