Background
Microbial secondary metabolites are low molecular weight compounds produced during the idiophase of bacterial growth. The Bacteria forms belonging to Pseudomonas, Bacillus and Streptomyces spp. Are prolific makers of secondary metabolites, which include peptides, polypeptides, phospholipids, pyrroles, phenazines, bacteriocins, lactones, anthracyclines, quinolones, aminoglycosides, macrolides, sisocoumarins, siderophores, and volatiles, among other naturally occurring substances. While these metabolites play important roles in human and animal health, they are not necessary for the growth and development of microorganisms like antibiotics, pigments, growth hormones, and anticancer agents (Ruiz et al., 2010).1 All these properties paved way for the use of secondary metabolites.
While validation results on microbial secondary metabolites show that the approaches yield values within acceptable tolerance levels, 2, 3, 4 our understanding of how to improve bioactive secondary metabolite production is currently limited. Most Bacillus species-produced molecules of biotechnological interest have been recognised thus far using lengthy screening methods and wide-ranging biochemical characterization. In general, continuous improvement in sequencing platforms has made sequencing technologies loftier and more powerful than ever before, and their application allows us to conduct investigations at a much deeper level and on a much bigger scale than ever before. Furthermore, bioinformatics evidence supports the idea that in-silico methodology may be used to predict new bioactive compounds from genome sequences, as well as their biochemical properties (Blin et al. 2017). 5
Novel Biosensor Design Using Synthetic Biology Applications
In the last few decades, it has become quite possible to develop resources and approaches to boost SM generation in bacteria. Numerous studies in synthetic biology have focused on the rebuilding of biosynthetic gene clusters in native or heterologous hosts, as well as the engineering of the host's regulatory network. These techniques have been widely employed on Gram-positive and Gram-negative bacteria and have now been utilised with non-model species as a result of the development of novel genetic tools. There has been a huge effort to construct new inducible systems in bacteria in order to design or restructure the regulatory networks that governs biosynthetic gene clusters. These elements can be utilised as biosensors to track the rate of generation of products of interest or as regulatory networks for gene pathway expression. (C. Liu et al.,2017).6 In this perspective, roughly some of the most important contemporary techniques, many of which are explored further below. The conventional method for increasing the production of biosynthetic gene clusters is to put these elements under the control of a powerful regulatory element, such as the T7 RNA polymerase/T7 promoter system, which is widely used for large-scale protein production in Escherichia coli (S. Tabor and C. C. Richardson, 1985). 7 Ross and colleagues used this strategy to produce a 34-kb gene cluster from Pseudoalteromonas piscicida in E. coli within the control of a T7 expression system (A. C. Ross et al., 2015). 8 This gene cluster is responsible for the production of an alterochromide lipopeptide. Because the T7-based expression system allows the introduction of an orthogonal expression device into a wide variety of host strains, such an approach does not necessitate the redesign of native regulatory elements from the original host (K. Terpe, 2006). 9 T7-based methods, for example, the one used to generate SM in Streptomyces in vitro, can also be easily adapted for cell-free conditions. (S. J. Moore et al., 2017)10 (D. Schwarz, F. Junge, F. Durst et al., 2007). 11, 10 Additionally, Strategies for assembling synthetic groups of genes could increase the production of useful compounds even further. Lack of effective genetic tools and induction measures, which are necessary for the circuit's success, is one of the most frequent constraints when it comes to developing a particular host. To increase SM production in Streptomyces venezuelae, Phelan and colleagues (2017) have released a series of new vectors for use in this organism. (R. M. Phelan.et al.,2017). 12
The genetic components required for fluorescence reporter systems, induction systems, and biological sensors in Rhodococcus opacus have been identified. (D. M. DeLorenzo et al., 2017). 13 In that study, utilising genome mining and transcriptome analysis, the scientists were able to improve inducible systems sensitive to arabinose (Pbad), anhydrotetracycline (Ptet), and acetamide (Pacet), as well as find endogenous expression systems responsible for compounds like phenol. (D. M. DeLorenzo et al.,2017). 13 Similar attempts for Streptomyces have been published (Y. Q. Sun et al., 2017 14; S. Li et al., 2018), 14 and genome mining combined with transcriptome analysis might seem to be an appealing methodology to recognize novel expression devices for use in synthetic biology initiatives designed in nonmodel species. To enable the construction of the requisite circuits, it is necessary in this scenario to provide sets of promoters with different strengths. With this target in mind, Yang and colleagues (2017) developed novel broadhost range promoters that could activate gene expression in Saccharomyces cerevisiae, Bacillus subtilis, and E. coli (S. Yang et al., 2017) 15. This groundbreaking research paved the way for the creation of universal synthetic clusters that could be tested in a variety of Gram-positive and Gram-negative hosts, as well as across life kingdoms. Rohlhill et al. (2017) 16 used FACS in conjunction with next-generation sequencing (NGS) to identify enhanced promoter variants produced by random mutagenesis of the E. coli formaldehyde inducible promoter, whereas Yang et al. explored only a small number of promoter variants using standard approaches. (J. Rohlhill et al., 2017). 16 By permitting researchers to examine brand-new promoters for SM-production engineering, the Sort-Seq method has the potential to enhance current capabilities. As an alternative, recent developments in computational methods have enabled the in silico creation of regulated promoters in E. coli (M. E. Guazzaroni et al., 2014 17; G. R. Amores et al., 2015). 18, 19
Microbial SM Production: Metagenomics as an Origin of Genetic Components
Using metagenomics to search the extensive metabolic variety present in the genomes of microorganisms found within living samples, researchers can collect the majority of the genetic material of bacteria resistant to growth. (L. Fernandez-Arrojo et al., 2010). 20 Unique functional gene clusters involved in the production of bioactive chemicals can be found by either cloning or sequencing DNA isolated from the microbial community occupants of a specific habitat (Figure 1) (M. E. Guazzaroni et al., 2010). 21 The creation of the activity-based metagenomic approach and subsequent screening of metagenomic libraries enables the determination of those specific genes encoding the required processes. (L. F. Alves et al., 2017). 22 Thus, large multienzyme clusters (such as nonribosomal peptide synthetases (NRPS), polyketide synthases (PKS), and terpene synthases, to name a few) could be functionally expressed or complete biosynthetic pathways could be recovered using large-insert libraries, which are typically built in cosmids or BACs (bacterial artificial chromosomes). (C. Hertweck.,2009). 23 According to J. Fu et al. (2012), 18 PKS type I and NRPS pathways are often structured into big assembly operons ranging in size from 20 to 100 kb. Numerous instances in the literature show how metagenomics has been successfully utilised to identify novel bioactive chemical pathways in a range of scenarios (J. J. Banik.,2010; 24 Z. Feng et al.,2011; 25 A. Felczykowska, H. A. Iqbal et al., 2014 26 27; S. A. Jackson et al.,2015; 28 C. J. Guo et al.,2017). Several studies employing next-generation sequencing technologies and subsequent bioinformatics mining of metagenomes have yielded novel gene clusters implicated in secondary metabolite synthesis. (F. Y. Chang.,2013; R. A. Cacho et al.,2014; 29 T. Weber et al.,2015; 30 K. Blin et al.,2017). 31
By adding new genetic elements (such as structural genes and regulatory sequences) to the synthetic biology toolkit, metagenomics has developed into a useful methodological tool for enhancing and expanding NP discovery from natural sources (Figure 2) (A. L. R. Santana-Pereira, C. A. Westmann, 2017). 32 Secondary metabolite genes are arranged into operons or clusters, which allows a collection of functionally related enzymes to synthesise compounds in a well-organized manner through a number of successive steps (P. Cimermancic et al., 2014). 33 The design and manufacture of innovative complex bioactive compounds would be enabled by novel combinations and rewiring of these enzymes, which carry out a wide range of biochemical alterations, as well as proper regulation of catalytic synergy. Freestone and colleagues (2017) 34 found a previously unidentified phosphonoacetic acid synthase by rerouting a gene cluster from Streptomyces sp. strain NRRL F-525 and using S. lividans as the expression host. (T. S. Freestone et al., 2017). 34 In parallel, algorithms that consider a number of genetic and nongenetic characteristics are being developed in an effort to optimise chemical biosynthesis (H. Zhou et al., 2015; 35 S. P. Bhatia et al., 2017) 36.
Figure 1
In order to increase and improve NP discovery using DNA isolated from the microbial populations that inhabit environmental materials, metagenomics has emerged as a crucial strategy. Novel genetic components are offered, which can be rewired to produce new complex bioactive substances. These components include structural genes, regulators, terminators, peptidesignals, transporters, and more.
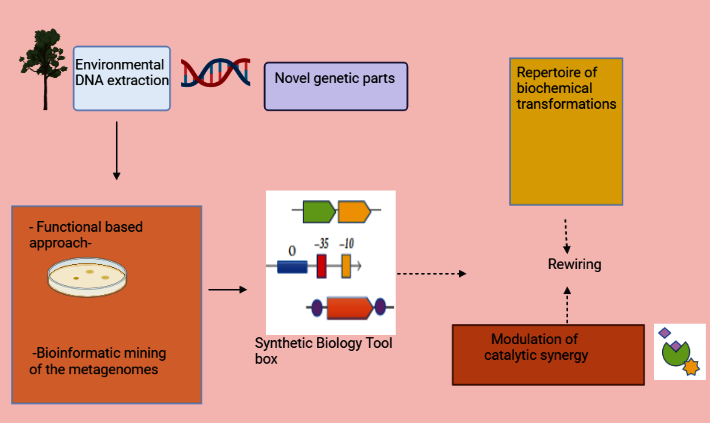
A key barrier to retrieving raw materials from metagenomes, on the other hand, is the use of E. coli as a host, a Gram-negative bacterium that is phylogenetically distinct from bacteria that are native makers of NPs. (M. E. Guazzaroni et al., 2015). 37 Thus, according to research by S. D. Bentley et al. (2002), 38 Y. Ohnishi et al. (2008), 39 and E. A. Barka et al. (2016) 40, Gram-positive bacteria from the phylum Actinobacteria are the most effective producers of a variety of secondary metabolites that are widely used in a number of medical applications. Some of the key restrictions that hinder E. coli from being employed as a host for NP discovery in metagenomic libraries are codon use, transporters, regulatory signals, proper protein folding, cofactor availability, and low substrate availability for secondary metabolite synthesis. (A. Lewin, 2017). A versatile model host for the heterologous expression of NPs was created by removing unnecessary genes from the genome of the industrial bacterium Streptomyces avermitilis (M. Komatsu et al., 2010).41 Actinobacteria have also developed a variety of genetic tools for genome editing and genetic modification. (including the well-known CRISPR/Cas system) (B. Gust et al.,2004; L. Du. et al.,2012; A. C. Jones et al.,2013;42 D. Du, H. Huang, R. E. Cobb, Y. Tong;2015).43, 44, 45 To summarise, while several limitations must be overcome before these bacteria can be used as potent host strains for metagenome library screening, current work, which includes synthetic biology techniques for framework engineering and genetic tool development, appears to be very promising.
Biochemical Engineering of Gram-Negative Microbials for the Generation of SMs
Several organizations have conducted research on the metabolic engineering of SM synthesis in bacteria in the last few years. In this sense, E. coli is one of the most well-studied Gram-negative bacteria, with various approaches for genetic manipulation and engineering discovered. This bacterium became an early adopter in the field of genetic engineering after generating the first genetically altered E. coli in 1973. (K. R. Choi et al., 2016). 46 A noteworthy instance is the utilization of E. coli co-cultures in the production of flavan-3-ols, a flavonoid subclass with several medical applications. This approach surpassed earlier attempts by a factor of 970 and allowed for the modification of a variety of variables including carbon supply, induction temperature, induction point, inoculation ratio, and strain selection (J. A. Jones et al., 2016). 47 In E. coli strains, genes involved in the manufacture of myrcene, an acyclic monoterpene, are also being affected. (E.M. Kim et al., 2015). Myrcene, a monoterpene molecule, has been utilised as a starting material for the production of more complex compounds in flavours, perfumes, cosmetics, vitamins, and pharmaceuticals (A. Behr et al., 2009). 48 Myrcene levels surged 34 times (58.19 12.13mg/L) after heterologous expression of the mevalonate (MVA) pathway (E.M. Kim et al., 2015). Furthermore, the US Department of Energy has designated 2-pyrrolidone, a glutamate derivate, as a crucial C4 "Top Value-Added Chemical from Biomass" (T. Werpy et al., 2004). 49 Because of its wide range of commercial applications, 2-pyrrolidone is a valuable chemical. It can be used to make nylon-4, which is more thermally stable and hydrophilic compared to its predecessors, using ring-opening polymerization. With this aim in mind, a team of scientists in California created a recombinant E. coli strain capable of generating 2-pyrrolidone utilising glutamate as a substrate. (S. J. Park et al., 2013) 50. To do this, in silico analysis was performed to identify two ORFs from type I PK gene clusters that were expected to be AMP-dependent synthetases. In recombinant E. coli expressing a glutamate decarboxylase and one of the synthetases, the efficiency of -pyrrolidone synthesis was increased by 25% (J. Zhang et al., 2016). 51 Wang et al. (2017) presented a coexpression approach in E. coli to produce trans-resveratrol. Resveratrol is a stilbene-family secondary metabolite that can be obtained from a variety of plants, aromatherapy products, and dietary substances (K. A. Roupe et al., 2006). 52 Stilbenes are used in humans to prevent cancer, heart disease, and neurological problems. (2006) (K. A. Roupe et al.). 52 Alongside E. coli, P. putida has been extensively used in metabolic engineering. P. putida is a Gram-negative bacteria that can metabolize a diverse spectrum of natural and synthesized chemical compounds, prompting various studies to investigate its potential use as a biocatalytic agent for industrial and ecological purposes. (V. A. P. Martins Dos Santos., 2004; 53 E. Mart´ınez-Garc´ıa., 2015). Taking this into account, Simm et al. (2016) reported a study in which they modified P. putida with two E. coli genes that produce active GGDEF and EAL domains, which are involved in c-di-GMP synthesis and deterioration, respectively (R. Simm., 2004). 54 P. putida mutations may influence biofilm formation in response to certain catalytic demands, such as the biodegradation of the environmental pollutant 1 chlorobutane. When the scientists added cyclohexanone to the growth conditions, the secondary metabolite produced by haloalkane in P. putida biofilm generating cells increased twice as much as in planktonic cells. (I. Benedetti et al., 2016). 55
Important research on the generation of beneficial secondary metabolites is included in the research topic. According to (Alenezi et al.2016), the biological activity of Aneurinibacillus migulans isolates was directly related to the synthesis of a novel gramicidin.
A collection of articles concentrates on the biosynthetic gene clusters that are involved in the fabrication of secondary metabolites in microorganisms. Another study (Rojas-Aedo et al.2016) described the role of the adr gene cluster in the manufacture of the powerful anticancer drug andrastin A in Penicillium roqueforti. Finally, (Nah et al. 2015) 56 evaluated the phylum Actinomycetes' potential for natural production (NP) via biosynthetic gene clusters (BGC) heterologous expression systems, as well as current methodologies generalised for substantial amount of NP BGCs in Streptomyces heterologous hosts.
Future Perspectives
Currently, it is critical to isolate novel and natural bioactive compounds to combat the failure of existing chemotherapeutic agents in treating infectious diseases due to increased drug resistance and a lack of knowledge of metabolites in the health industry. The study articles compiled for recent development and Technological Challenges investigate the role of microorganisms from various sources, demonstrating varied biological activities. The development of methodologies to understand the detailed mechanisms of cryptic genes and their relationship to the generation of bioactive chemicals is a key challenge that must be addressed. We must also place a greater emphasis on the co-culture of diverse microorganisms that have a good synergistic effect in order to develop novel bioactive secondary compounds.
