Introduction
Eighty percent of medicinal herbs have already been shown to be non-toxic, making them safer to use regularly than traditional antibiotics.1 Recent decades have seen the emergence of innovative antibacterial drugs derived from plants due to their abundance of active phytochemicals with antimicrobial action.2, 3 In the traditional system, Coccinia grandis is utilized in cooking and has been described as an ethnomedicinal plant with various pharmacological qualities.4 Many common uses include treating diabetes, leprosy, jaundice, asthma, bronchitis, burns, skin eruptions, tongue sores, earache, indigestion, eye infections, nausea, insect bites, and fever.5
The increasing prevalence of bacteria resistant to therapeutically important antimicrobials is causing new generations of antibiotics and empirical medications to become less effective against infectious disorders.6 However, some strains of carbapenem resistance adhering to metagenomic sequencing were discovered during the bacteriological evaluation of patients presenting with deep neck space infection based on the severity and clinical factors.7 It was found that the most common causes of deep neck infections in the head and neck area, as well as the failure of antibiotic treatment, include dental caries, trauma, airway obstruction, oropharyngeal infection, severe ear infection, and neck abscess. However, the risky condition usually worsens over time, particularly when deep neck infections are the source of malignancy.
Research on possible new drugs and antimicrobial components made from various natural sources has been going on to face it.8, 9 Consequently, some antibiotic exposure has been associated with toxicity and frequent clinical failure.10 Plant-based medications are now desperately needed to combat the current drug resistance issue. Drug discovery has also advanced significantly in recent years with the help of molecular modeling, and computational biology. However, compounds generated from natural products continue to be a significant source of medications for human use.11 Validating the unattributed biological activities of drugs and targets has been made easier with the use of in-silico techniques. Computational screening seems to be a substantial method of saving time, money, and human resources as compared to the traditional drug development strategy. Furthermore, it has predicted physicochemical properties from molecular and structural features of several chemical compounds and verified binding affinities to targets.12, 13
The present study emphasizes on the in-silico inhibitory actions of phytocompounds of C. grandis through molecular docking analysis. Moreover, the antimicrobial investigation of Klebsiella pneumoniae, Staphylococcus aureus, and Candida albicans was explored.
Materials and Methods
Study subjects
The head and neck infected swab samples were collected from various purulent discharge sites of patients who were hospitalized at the Institute of Medical Sciences and SUM Hospital's Otorhinolaryngology Department in Bhubaneswar, Khurda, Odisha, India, between 2020 and 2022. The subjects or their legal guardians gave their informed consent before the swab samples were collected.
Inclusion and exclusion criteria
The samples were taken from the HNI sites using sterile swabs, and they were processed appropriately. The study included patients with both scanty and profuse purulent discharge, while those with no purulent discharge from the infection sites and only mild swelling and redness were not included. In this investigation, both bacterial and candidal isolates were included.
Isolation, identification, and antimicrobial susceptibility profiling of collected samples
Purulent discharged swab samples were aseptically collected, streaked on Blood Agar (BA), MacConkey Agar (MAC), and Sabouraud dextrose Agar (SDA), and then incubated overnight at 37ºC. Swabs were also incubated overnight with 2 ml of nutrient broth (NB) for short-term storage. Standard protocols were followed for antibiotic profiling, Vitek 2, and preliminary biochemical identification. 14, 15, 16 Several antibiotics were used in the Vitek 2 approach to investigate the antimicrobial susceptibility pattern. Gram-negative bacteria were treated with Ticarcillin, Piperacillin, Ceftazidime, Cefepime, Aztreonam, Doripenem, Meropenem, Amikacin, Gentamycin, Ciprofloxacin, Levofloxacin, tetracyclin, and Colistin, while gram-positive bacteria were treated with Cefixime, Penicillin, Oxacillin, Amoxicillin, Gentamycin, Ciprofloxacin, Levofloxacin, erythromycin, Clindamycin, Linezolid, Daptomycin, Teicoplanin, Vancomycin, Tetracycline, and Tigecycline. The antifungals used against the candidal isolate were fluconazole, voriconazole, caspofungin, micafungin, amphotericin B, and flucytosine.
Protein preparation
The target proteins used in this study were obtained from the literature study and the database search were used as target receptors for the phytocompounds, which act as ligands. The proteins of this investigation were expressed in their respective microorganisms such as; Klebsiella pneumoniae:6LHE,17 Staphylococcus aureus:5M18,18 Candida albicans:5FSA.19 The quaternary structure of 6LHE (Gold-bound NDM-1), 5M18 (PBP2a from MRSA, and 5FSA (14-alpha demethylase CYP51) was downloaded and obtained from the SWISS Model (https://swissmodel.expasy.org/)20 and prepared using Discovery Studio software.
Ligand preparation
The phytocompounds of this investigation were retrieved from kinds of literature stating the pharmaceutical properties of C. grandis phytocompounds. 21, 22 Phytochemicals were retrieved from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/) and optimized using Avogadro software (https://prankweb.cz/).
Molecular docking study
The molecular docking of the protein and ligands was performed to study the stabilizations of the phytocompounds as ligands so that they can be utilized as therapeutic agents such as antimicrobial agents to combat antimicrobial resistance. The Molecular docking was conducted with AutoDock 4.2 (https://autodock.scripps.edu/download-autodock4/), and the binding pocket was identified through the Prank Web server.
Results
Microorganisms
The bacterial and candidal isolates used in this study were identified with the molecular identification procedure using universal primers of the respective pathogens. 16 The bacterial and candidal isolates used in this investigation were, Klebsiella pneumoniae, Staphylococcus aureus and Candida albicans and the genebank accession numbers of the pathogens were OP889575, OP889672, OR345133 respectively.
Antibiotic susceptibility pattern of the microorganisms
The Vitek 2 antimicrobial susceptibility patterns of K. pneumoniae, S. aureus, and C. albicans were presented in Table 1. The antibiotics and the antifungals used against the microorganisms revealed that most of antimicrobials were resistant to the pathogens.
Table 1
Vitek 2 antimicrobial profiling of the microorganisms
[i] *TI-Ticarcillin, PI-Piperacillin, CAZ- Ceftazidime, CPM-Cefepime, AT- Aztreonam, DOR- Doripenem, MOR- Meropenem, AK- Amikacin, GEN- Gentamycin, CIP-Ciprofloxacin, LE- Levofloxacin, TE-Tetracyclin, CL-Colistin.*CX-Cefixime, P-Penicillin, OX-Oxacillin, AMX-Amoxicillin, GEN-Gentamycin, CIP-Ciprofloxacin, LE-Levofloxacin, MO-Moxifloxacin, E-Erythromycin, CD-Clindamycin, LZ-Linezolid, DAP-Daptomycin, TEI-Teicoplanin, VA-Vancomycin, TE-Tetracycline, TGC-Tigecycline
Molecular docking analysis
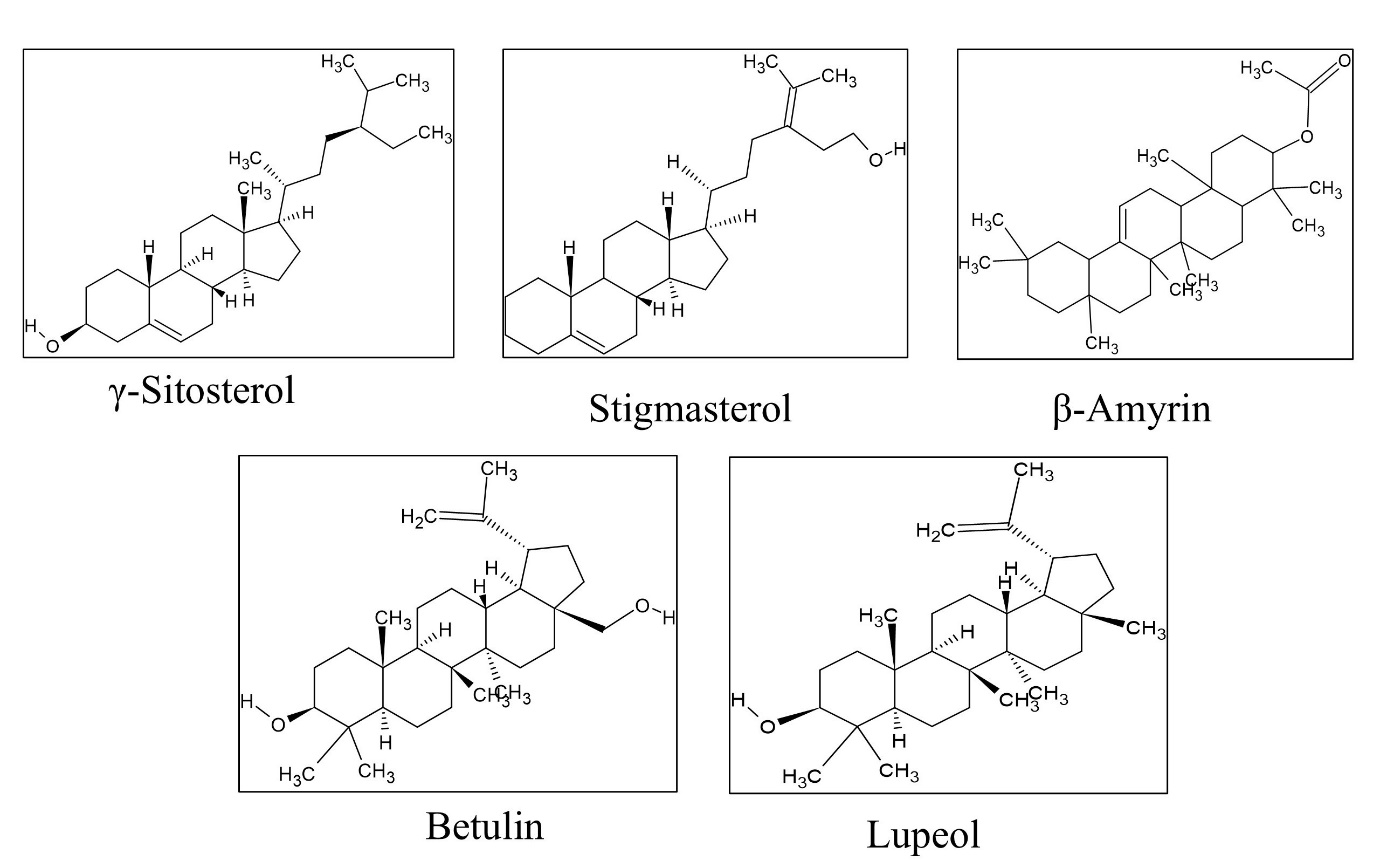
In this study, we investigated the binding characteristics of γ-Sitosterol, Stigmasterol, β-Amyrin, Betulin, and Lupeol. The optimized PubChem versions of all ligands are presented in Figure 1. These FDA-approved phytocompounds were complexed with the protein targets of respective pathogens. Through molecular docking analysis, it was observed that CID: 259846, 66825540, 259846 (Lupeol, γ-Sitosterol, β-Amyrin) exhibited the highest binding energy among the complexes, with a value of −10.18 kcal/mol, −8.32 kcal/mol and −8.76 kcal/mol in 5FSA, 5M18 and 6LHE respectively (Table 2). However, for their large size and strained cylindrical structure resulted in a limited number of hydrogen bonds within the active cavity but still, some compounds showed hydrogen bonds in the pockets (Figure 2, Figure 3, Figure 4).
The molecular docking analysis of phytocompounds with 5M18 (S. aureus) revealed the best binding score with γ-Sitosterol of binding energy -8.32Kcal/mol followed by lupeol with -7.92Kcal/mol and the interacting amino acids of the highest binding affinity contain compound were TyrA:470, LeuA:260, LysA:254, ProA:471, LeuA:365 (Table 2, Figure 2).
Figure 2
Molecular docking analysis of phytocompounds with protein 5M18 expressed in S. aureus; A. γ-Sitosterol, B. Stigmasterol, C. β-Amyrin, D. Betulin, E. Lupeol
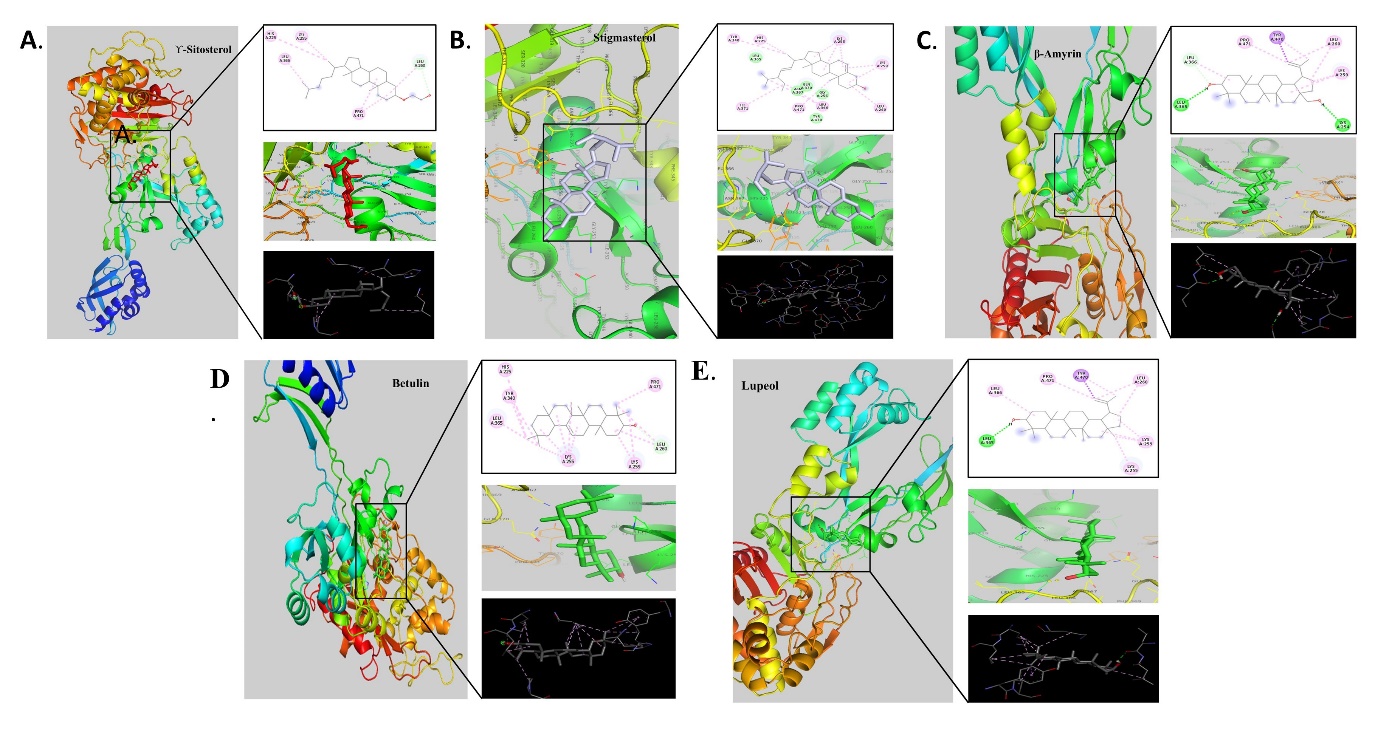
The molecular docking analysis of phytocompounds with 6LHE (K. pneumoniae) revealed the best binding score with β-Amyrin of binding energy -8.76 Kcal/mol followed by lupeol with -8.68 Kcal/mol and the interacting amino acids of the highest binding affinity contain compound were HisA:161, CysA:180, TrpA:65, LysA:183, ValA:45, HisA:222 (Table 2, Figure 3).
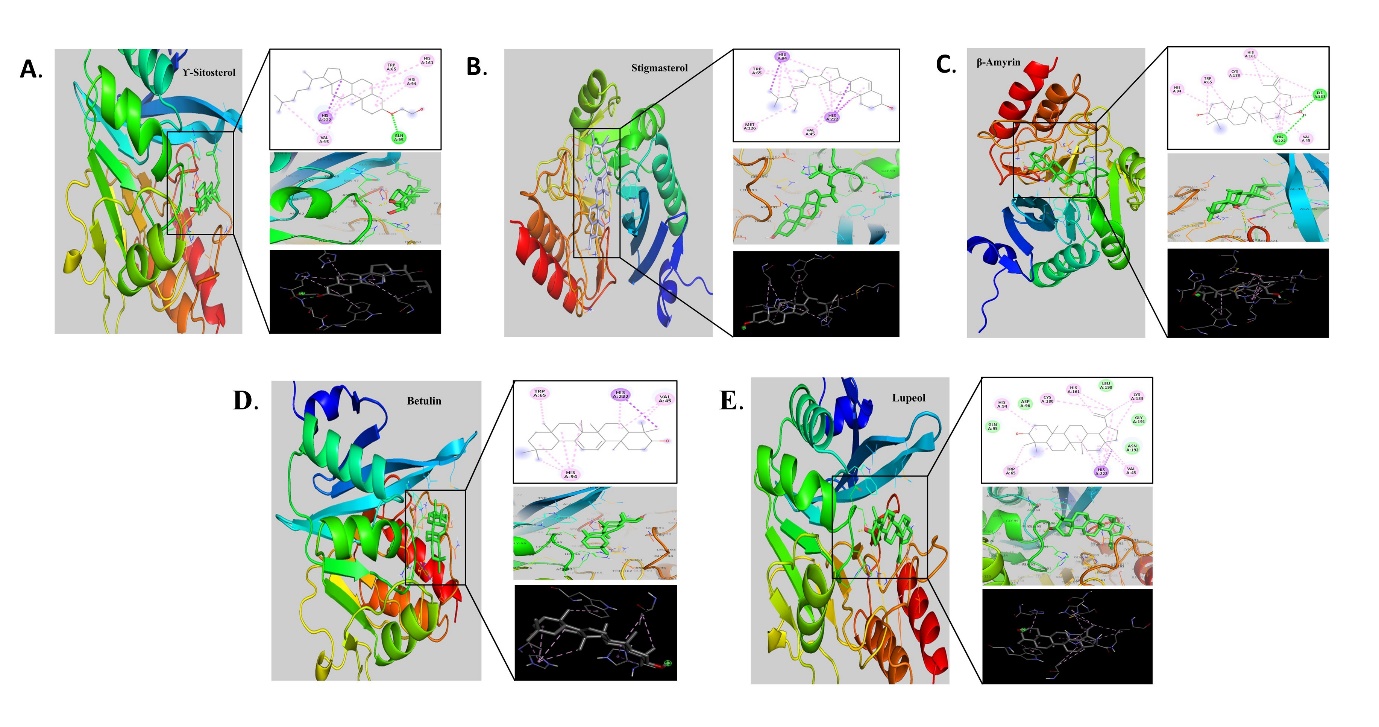
Figure 3
Molecular docking analysis of phytocompounds with protein6LHE expressed in K. pneumoniae; A. γ-Sitosterol, B. Stigmasterol, C. β-Amyrin, D. Betulin, E. Lupeol
The molecular docking analysis of phytocompounds with 5FSA (C. albicans) revealed the best binding score with stigmasterol of binding energy -10.54 Kcal/mol followed by lupeol with -10.18 Kcal/mol and the interacting amino acids of the highest binding affinity contain compound were IleA:379, MetA:374, ProA:375, LeuA:376, AlaA:476, GlyA:308, GlnA:479, CysA:470, PheA:463
Figure 4
Molecular docking analysis of phytocompounds with protein 5FSA expressed in C. albicans; A: γ-Sitosterol, B: Stigmasterol, C: β-Amyrin, D: Betulin, E. Lupeol
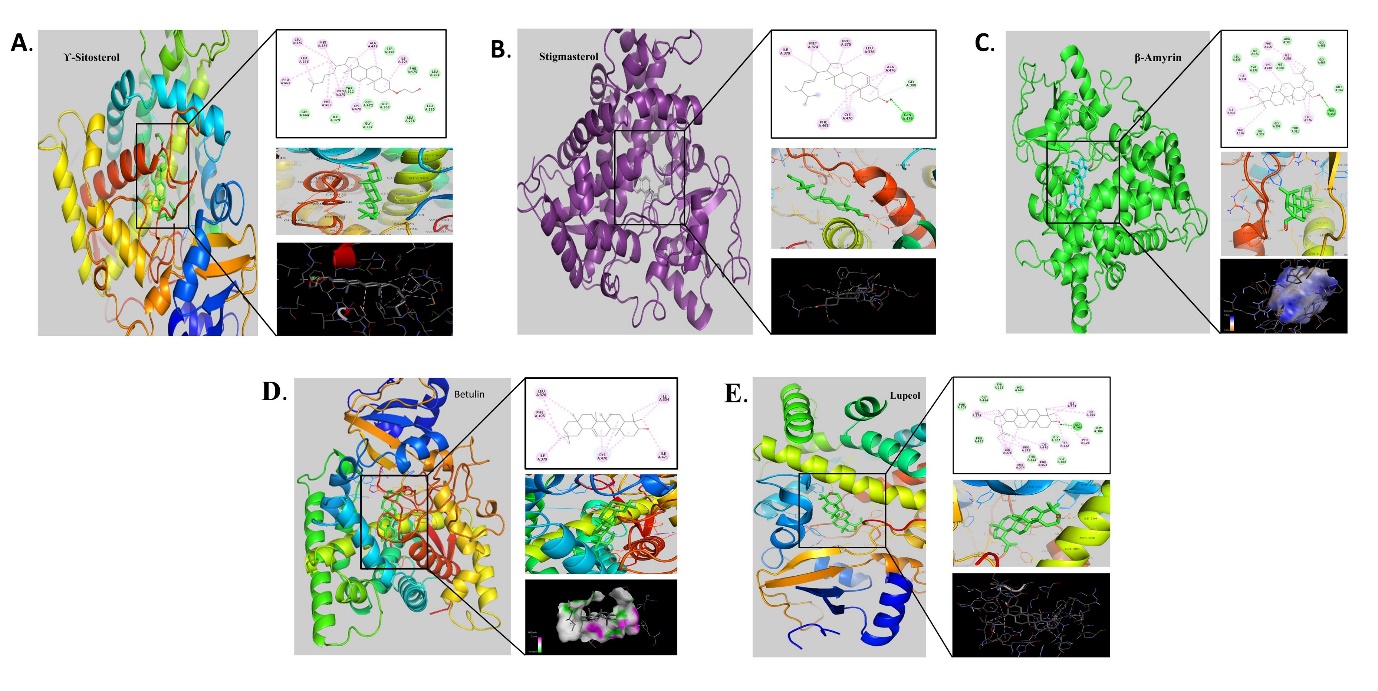
Table 2
Binding scores and interacting amino acids of phytocompounds with 5M18, 6LHEand 5FSA receptor protein
Discussions
K. pneumoniae (OP889580), the ꞵ-lactam-resistant strain employed in this investigation, was isolated from the head and neck region. Due to its extensive dissemination, the multidrug-resistant bacteria K. pneumoniae has been on the rise over the last 20 years. Furthermore, it is alarming how quickly carbapenem-resistant K. pneumoniae is emerging. 23 The another strain of interest in our investigation, methicillin-resistant S.aureus has emerged and disseminated globally since the 1960s and has become the leading cause of bacterial infectious diseases. 24 Regarding the burden of fungal infection, azole-resistant antifungals remain a multicentric concern in treating Candida infection. However, Fluconazole is mostly preferred antifungal against Candida infections, as they exhibit low toxicity and is inexpensive compared to other antifungals. 25 The complete identification procedure, antimicrobial resistance, and the population genetic parameters of these bacteria were discussed earlier in our preliminary study. 16
In our investigation phytocompounds of C. grandis played the key role as a therapeutic agent against head and neck infection pathogens. However, numerous phytocompounds have already been evaluated from C. grandis, 26, 27 each of which has a variety of health benefits. 28 The phytocompounds examined in this study were similar to the pharmaceutically significant phytocompounds identified in the methanolic and aqueous extract of C. grandis fruit, which included polyphenols, flavonoids, terpenoids, triterpenoids, alkaloids, and phenolic compounds. 29 Although the phytocompounds of C. grandis may differ throughout studies, the majority of them share some key phytocompounds. 21, 22
In-silico evaluation of compounds against the target proteins or genes can define the possible therapeutic actions without any confusion and effort. Due to less expenditure, easy handling, less manpower, and no chemical investment, the computational approach of drug development is proved to be more validated before stepping into the in-vitro laboratory activities. 30, 31 Therefore, this study emphasized molecular docking analysis of the phytocompounds of C. grandis to check their stability against the targeted proteins of some pathogens isolated from head and neck infection. The analysis revealed excellent binding affinities with all phytocompounds against their respective proteins and stated their antimicrobial property to combat multidrug resistance. A study conducted in Bangladesh by Ahmed et al., (2024), 32 implemented the same approach toward the therapeutic finding of diabetes. The phytocompounds, γ-Sitosterol, Stigmasterol, and β-Amyrin were shown to be the best binding affinities with the three target proteins. Among them, stigmasterol with a binding affinity of -10.54 Kcal/mol, was foot forward with the highest binding affinity against the 5FSA target protein of C. albicans. In recent, one review conducted by Bakrim et al., (2022), 33 was found regarding the health benefits of stigmasterol, but the confirmation of therapeutic potential with the binding affinity was evaluated in this investigation. Subsequently, here is another interesting thing found that, following the best binding affinities with these three phytocompounds, lupeol was one of them having the second most binding affinities. In addition to that, it was seen to be the most distinctive phytocompounds among all as the target proteins of both bacteria and Candida strain were shown to have a good binding affinity with it. There are several studies regarding the antibacterial 34 and antifungal 35 activities of lupeol individually, but there are lack of studies stating lupeol as a potent phytocompound that can inhibit both bacteria and Candida infections isolated from a plant (C. grandis), which was used as culinary purpose in most Asian countries.
However, there is no further study reported about the proper mechanism regarding the antimicrobial activity of these phytocompounds yet. However, extensive research on phytocompound derivatives is still in the preliminary stage; hence, more in-depth studies are essential to isolate the pure compounds as therapeutic approaches for the future practical applications of these natural products.
Conclusion
The concluding remarks of the current study are that γ-Sitosterol, Stigmasterol, β-Amyrin, and lupeol showed good binding affinities with the target proteins of the pathogens. Concerning the antimicrobial resistance of the standard antimicrobials used earlier against the pathogens, the necessity of a natural antimicrobial agent finding became essential. This study revealed the computational validity of the phytocompounds against the target proteins of the pathogens, which would be the next generation of therapeutic steps regarding infectious diseases. In addition to the computational validation, there is surely required further isolation, in-vitro as well as in-vivo validation of these natural compounds in the laboratory setup to lighten future therapeutics.