- Visibility 5 Views
- Downloads 2 Downloads
- DOI 10.18231/j.ijmmtd.2021.034
-
CrossMark
- Citation
Biofilm production and antibiotic resistance pattern among bacterial isolates from wound samples in a rural tertiary care teaching hospital
- Author Details:
-
Venkatesha D
-
Dhanalakshmi T. A
-
Shakthi R *
Introduction
Pyogenic wound infections are the one of the leading cause of morbidity and mortality worldwide.[1], [2] Wound sepsis rate in India is around 10-33%. Infections of the wound also contribute huge economic burden because of prolonged hospital stay.[3] Upsurge of antimicrobial resistance has provided a new angle to the current problem of wound infections.[4] Biofilms have enormous negative impact on health care system.[5] Formation by biofilms by wound isolates impair healing of the wound, reduce host immune response and further add on to the development of antibiotic resistance as biofilms impede delivery of antibiotics, cause degradation of antibiotics, promote horizontal transfer of resistance genes [1].Biofilms complicate patient treatment.[6] Data on biofilm formation by isolates is required to modify treatment modality and the outcome of clinical condition. [7] The spectrum of bacteria causing wound infections and their antibiotic susceptibility patterns exhibit geographic variability and also changing trends noted with respect to time.[8], [9] Updated information regarding the bacteriological profile and their antibiogram is valuable for implementing strategies towards empiric treatment of wound infections, in adopting efficient control protocols and in formulation of suitable antibiotic policies for treatment of wound infections in the region.[6], [8] With this background the present study was carried out to determine the bacteriological spectrum of wound infections and their antibiogram to commonly used antibiotics and to detect the biofilm production by the isolates.
Materials and Methods
The present cross sectional study was carried out in the department of Microbiology, Adichunchanagiri institute of Medical sciences from September 2016 to August 2017. Two hundred and forty pus samples from various wounds were collected and submitted to microbiology laboratory of the hospital and were processed as per standard procedures.[10] Both inpatients and outpatients were included in the study.
Repeated isolates from the same patient and patients who were on antibiotic therapy or had history of antibiotic intake within one week prior to sample collection and anaerobic isolates were excluded from the study. Informed consent from patients and ethical clearance from the institution were obtained for the study.
Specimens were inoculated on to Blood agar and MacConkey agar plates (procured from HiMedia Mumbai India) and incubated for 48 hours at 370C under aerobic conditions. Isolates were identified by standard microbiological methods.[10] Antibiotic susceptibility testing was carried out by Kirby Bauer disc diffusion method as per Clinical Laboratory Standards Institute guidelines with ATCC Staphylococcus aureus ATTCC 25923 and Escherichia coli ATCC 25922 as control strains.[11], [12]
For biofilm detection, S.epidermidis ATCC 35984 and S.epidermidis ATCC 12228 were used as positive and negative controls respectively. [13]
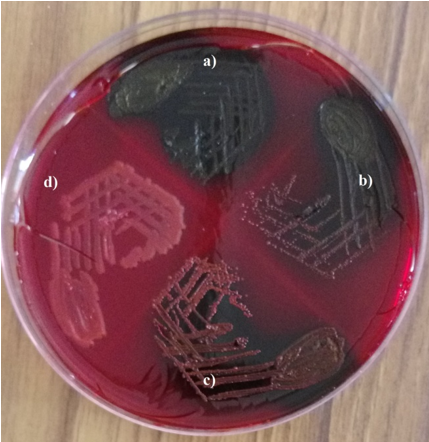
Isolates were tested for biofilm production by Congo red agar method. Isolates were inoculated onto Congo red agar plate (procured from HiMedia Mumbai, India) and were incubated for48 hours at 370C under aerobic conditions. The appearance of black colonies with dry crystalline consistency was taken as positive for slime production. Isolates producing very dark coloured colonies were interpreted as strong biofilm producers. Those bacteria forming black colonies were considered as moderate biofilm producers and those producing almost black colonies were noted as weak biofilm producers. Isolates forming red colonies were considered as non-biofilm producers. Each test was interpreted by two different observers[14], [15] ([Figure 1]).
Statistical analysis
Statistical analysis was done using Microsoft excel. The data analysis involved transcription, preliminary data inspection, content analysis and interpretation. Percentages were used in this study to analyse variables
Results
Out of 240 pus isolates studied, Staphylococcus species were the most commonly isolated bacteria (48.85) followed by Pseudomonas species (11.7%) ([Table 1]).
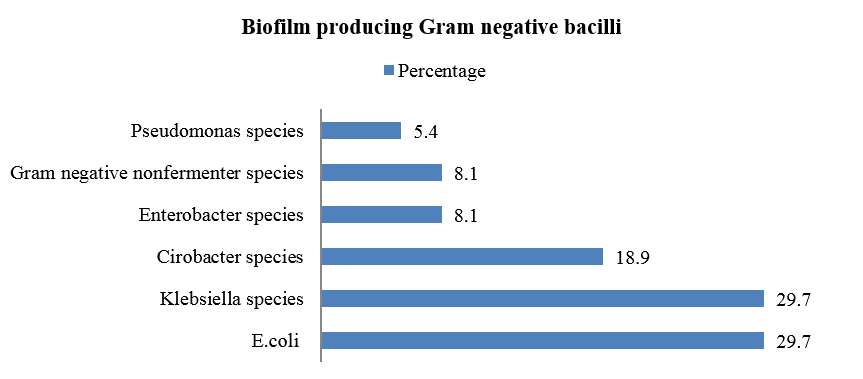
Biofilm was produced by 49.2% of total isolates. Among biofilm producers 39% were strong biofilm producers followed by mild (37.3%) and moderate biofilm producers (23.7%). Among Gram negative bacilli isolated, biofilm production was observed more among E. coli and Klebsiella species (29.7% each) followed by Citrobacter species (18.9%).([Figure 2]).
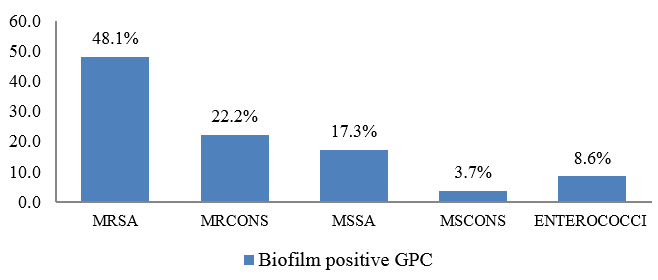
Out of total Gram positive cocci isolated in the study, majority of biofilm production was seen among Methicillin Resistant Staphylococcus aureus (MRSA) isolates (48.1%) followed by Methicillin Resistant Coagulase negative Staphylococci (22.2%). ([Figure 3])
Majority of Gram negative bacilli were found sensitive to Colistin, Tigecycline, and aminoglycosides followed by Imipenem. ([Table 2])
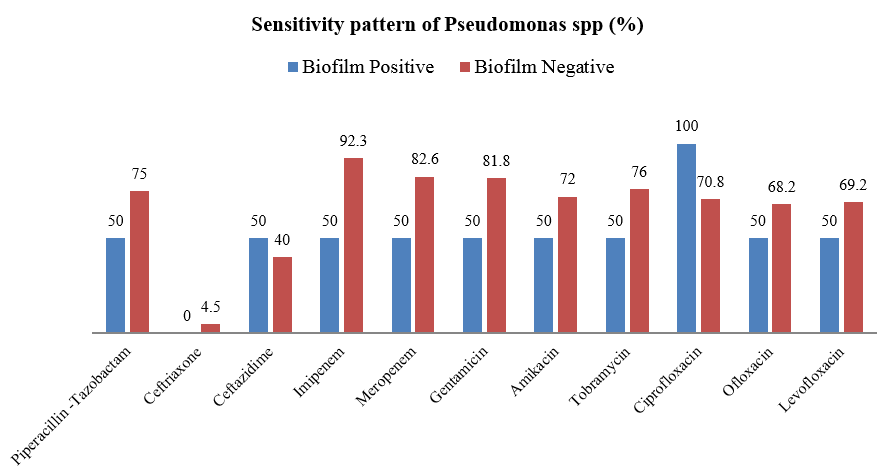
Majority of Gram positive cocci were sensitive to Teicoplanin, Vancomycin and Linezolid ([Table 3]). Majority of Pseudomonas species were sensitive to ciprofloxacin followed by carbapenems and aminoglycosides. ([Figure 4])
|
Bacteria isolates |
Number (%) |
|
Staphylococci species |
117 (48.8) |
|
Pseudomonas species |
28 (11.7) |
|
Klebsiella species |
24 (10.0) |
|
E.coli |
21(8.8) |
|
Citrobacter species |
17(7.1) |
|
Gram negative non-fermenters other than Pseudomonas and Acinetobacter spp |
10 (4.2) |
|
Enterococcus species |
9 (3.8) |
|
Enterobacter species |
4 (1.7) |
|
Proteus species |
3 (1.3) |
|
Streptococcus species |
2 (0.8) |
|
Acinetobacter species |
2 (0.8) |
|
Morganella species |
2 (0.8) |
|
Providencia species |
1 (0.4) |
|
Total |
240 (100) |
|
Antibiotics |
Biofilm positive |
Biofilm Negative |
|
Ampicillin |
2.7 |
1.4 |
|
Amoxicillin -Clavulanic acid |
2.9 |
2.8 |
|
Piperacillin-Tazobactam |
37.8 |
56.5 |
|
Ceftriaxone |
35.3 |
13.8 |
|
Ceftazidime |
29.7 |
32.4 |
|
Cefepime |
44.4 |
43.1 |
|
Imipenem |
50 |
72.6 |
|
Ciprofloxacin |
38.2 |
49.3 |
|
Ofloxacin |
40.6 |
53.7 |
|
Gentamicin |
58.3 |
67.2 |
|
Amikacin |
67.6 |
65.3 |
|
Tobramycin |
63.9 |
77.5 |
|
Tigecycline |
75 |
66.7 |
|
Colistin |
100 |
100 |
|
Antibiotics |
Biofilm Positive |
Biofilm Negative |
|
Penicillin |
6.4 |
13.6 |
|
Amoxicillin-Clavulanic acid |
11.8 |
20.9 |
|
Ceftriaxone |
0 |
0 |
|
Cefotaxime |
17.2 |
27.9 |
|
Erythromycin |
22.2 |
32.6 |
|
Clindamycin |
45.2 |
53.5 |
|
Ciprofloxacin |
13.7 |
29.5 |
|
Ofloxacin |
7.4 |
36.7 |
|
Gentamicin |
41.7 |
55.6 |
|
Amikacin |
50.6 |
62.2 |
|
Tetracycline |
78.4 |
73.3 |
|
Chloramphenicol |
69.6 |
71.8 |
|
Cotrimoxazole |
38.5 |
43.2 |
|
Vancomycin |
100 |
100 |
|
Linezolid |
98.8 |
97.8 |
|
Teicoplanin |
100 |
100 |
Discussion
Wound infection is one of the most common and serious complication among the hospital acquired infections. Wound infection can increase the length of hospital stay and accounts for mortality rate up to 70-80%.[12]
In the present study, a dominance of Gram Positive bacteria as the causative agent of pyogenic lesions was seen similar to studies by Tiwari et al., Lee et al., 2009.[16], [17] This is in contrast to other studies where Gram negative bacteria are most common pathogens isolated.[12], [18] Staphylococcus forms the normal microbial flora of the skin and anterior nares and hence can easily contaminate the wound. [2], [5], [6]
Staphylococcus aureus (48.85%) was the most common Gram positive isolate which similar to the various studies.[12], [16], [17] The prevalence of MRSA (39%) reported in our study is in accordance with the studies of Kshetry et al,.(37.6%), Sanjana et al.,(37.6%), Dibah et al.,(46.3%).[19], [20], [21] However, lower rates were reported by Rozina AK et al., Tiwari et al., Lee et al.,[12], [16], [17]. The difference in the rates of isolation of MRSA might be due to the difference in the level of irrational antibiotic use, level of hygienic condition maintained in different hospitals and effective implementation of hand hygiene program.[12]
Among Gram Negative bacilli, the most common isolate was Pseudomonas species (11.7%) followed by Klebsiella species (10%) and Escherichia coli (8.8%). This is in accordance with the study of Mukhopadhyay et al,.[22] but is in contrast with the study of Pal K et al., where Escherichia coli was the most common Gram negative isolate.[23] This inconsistency in bacterial pattern may be due to regional variation of bacterial profile, habits of the local people and also due to the fact that all the Indian studies done so far involve community set up and were not centered on hospitals.[24]
Antimicrobial resistance is an innate feature of bacterial biofilm that, in addition to the increasing resistance among clinical strains, may complicate patient treatment.[6] In this study biofilm was produced by 49.2% of total isolates. Out of total Gram positive cocci isolated in the study, majority of biofilm production was seen among Methicillin Resistant Staphylococcus aureus isolates (48.1%), which is similar to the studies of Shrestha et al (44.9%) and Ansari et al (43.1%).[25], [26]
Majority of Gram negative bacilli were found sensitive to Colistin, Tigecycline, and aminoglycosides followed by Imipenem which is similar to various studies.[8], [9], [27] Majority of Pseudomonas species were sensitive to ciprofloxacin followed by carbapenems and aminoglycosides, this is in accordance with Sukumar N, et al.,; Sharma V et al., and Amabegaum B et al.,[8], [9], [28]
Majority of Gram positive cocci were sensitive to Teicoplanin, Vancomycin and Linezolid which is in agreement with other studies. [1], [9], [29], [30]
The prevalence and pattern of Antimicrobial resistance among wound isolates show variability according to geographic location, endemicity of pathogen in the locality and climate conditions.
Limitation of the Study
In this study, clinical correlation was not done. Small sample size did not allow us to conduct advanced statistical analysis, which could have potentially strengthened this study
Conclusion
In the present study, pyogenic wound infections were mainly caused by S. aureus, Pseudomonas spp, Klebsiella spp., and Escherichia coli. Both Gram positive and Gram negative bacteria with biofilm production showed modest increase in resistance to few antibiotics compared to those without biofilm production.
Continuous surveillance is necessary to update the knowledge of antimicrobial susceptibility pattern of clinical isolates to provide the appropriate dose regimen and treatment schedule against pyogenic wound infections and to limit the expanding threat of drug resistance. Early identification and adopting efficient control protocol against biofilm forming organism can prevent the most serious nosocomial infections and improve the outcome of the condition.
Conflict of Interest
The authors declare that there are no conflicts of interest in this paper.
Source of Funding
None
References
- B P Rijal, D Satyal, N P Parajuli. High burden of antimicrobial resistance among bacteria causing pyogenic wound infections at a tertiary care hospital in Kathmandu, Nepal. J Pathog 2017. [Google Scholar] [Crossref]
- N A Kassam, D J Damian, D Kajeguka, B Nyombi. Spectrum and antibiogram of bacteria isolated from patients presenting with infected wounds in a tertiary Hospital, northern Tanzania. BMC Res Notes 2017. [Google Scholar] [Crossref]
- N Sukumar, S Rajesh. Aerobic bacterial isolates and their antibiotic susceptibility pattern from pus samples in a tertiary care Government Hospital in Tamil Nadu, India. Int J currMicrobiol App Sci 2017. [Google Scholar]
- A Biradar, F Farooqui, R Prakash, SY Khaqri, I Itag. Aerobic bacteriological profile with antibiogram of pus isolates. Indian J Microbiol Res 2016. [Google Scholar] [Crossref]
- D C Kaur, Wankhede. Biofilm formation and antimicrobial susceptibility pattern of Methicillin Resistant Staphylococcus aureus from wound infection. Asian Pc J Health Sci 2014. [Google Scholar]
- DC Kaur, AS Khare. Biofilm formation and antibiotic susceptibility pattern in MRSA strains in a tertiary care rural hospital. Indian J Basic Appl Med Res 2013. [Google Scholar]
- S Asati, U Chaudhary. Prevalence of biofilm producing aerobic bacterial isolates in burn wound infections at a tertiary care hospital in northern India. Ann Burns Fire Disasters 2017. [Google Scholar]
- R Trojan, L Razdan, N Singh. Antibiotic Susceptibility Patterns of Bacterial isolates from Pus samples in a Tertiary Care Hospital of. Int J Microbiol 2016. [Google Scholar] [Crossref]
- V Sharma, G Parihar, V Sharma, H Sharma. A study of various isolates from Pus sample with their antibiogram from Jin Hospital, Ajmer. J Dent Med Sci 2015. [Google Scholar]
- JG Collee, TJ Mackie, JE McCartney, A Simmons. Mackie & McCartney practical medical microbiology. 14th Edn. 1999. [Google Scholar]
- . Clinical and Laboratory Standards. PerformanceStandards for Antimicrobial Susceptibility testing; Twenty fifth Informational Supplement. CLSI Document M100-S25.Wayne PA, USA: Clinical and Laboratory Standards Institute. 2015. [Google Scholar]
- A K Rozina, J Mahwish, K Mohammed. Bacteriological profile and antibiogram of isolates from pus samples in a tertiary care centre. Int J Curr Microbiol App Sci 2018. [Google Scholar]
- S Niveditha, S Pramodhini, S Umadevi, S Kumar, S Stephen. The isolation and the biofilm formation of uropathogens in the patients with catheter associated urinary tract infections. J Clin Diagn Res 2012. [Google Scholar]
- K Murugan, M Usha, AS Al-Sohaibani, M Chandrasekaran. Biofilm forming conjunctivitis multi drug resistant Staphylococcus spp among the ocular patients. Pol J Microbiol 2010. [Google Scholar]
- AA Ferreira, PAC Tette, RCS Mendonca, ASC Soares, MM Carvalho. Detection of exopolysaccharide production and biofilm related genes in Staphylococucus species isolated from poultry processing plant. Food Sci Technol 2014. [Google Scholar]
- HK Tiwari, AK Das, D Sapkota, VK Pahwa, K Sivarajan. Methicillin resistant Staphylococcus aureus: prevalence and antibiogram in a tertiary care hospital in western Nepal. J Infect Dev Ctries 2009. [Google Scholar]
- CY Lee, PY Chen, FL Huang, F Lin. Microbiologic spectrum and susceptibility pattern of clinical isolates from the pediatric intensive care unit in ingle medical centre-6years experience. J Microbiol Immunol Infect 2009. [Google Scholar]
- M Zubair, A Malik, J Ahmad. Clinico-microbiological study and antimicrobial drug resistance profile of diabetic foot infections in North India. Foot 2011. [Google Scholar]
- AO Kshetry, ND Pant, S Bhandarirkhatri, KL Shrestha, SK Upadhaya. Minimum inhibitory cocncentration of Vancomycin to methicillin resistant Staphylococcus aureus isolated from different clinical samples at a tertiary care hospital in Nepal. Antimicobial Resist Infect Control 2016. [Google Scholar] [Crossref]
- R Sanjana, R Shah, N Chaudhary, Y Singh. Prevalence and antimicrobial susceptibility pattern of Methicillin resistant Staphylococcus aureus(MRSA) in CMS- teaching hospital: a preliminary report. J Coll Med Sci-Nepal 2010. [Google Scholar]
- S Dibah, M Arzanlou, E Jannati, R Shapouri. Prevalence and antimicrobial resistance pattern of Methicillin resistant Staphylococcus aureus (MRSA) strains isolated from clinical specimens in Ardabil, Iran. Iranian J Microbiol 2014. [Google Scholar]
- M Mukhopadhyay, S Podder, S Bhattacharya. Microbial contamination of Indian currency notes and coins in Kolkata, West Bengal - A Survey. Int J Sci Res 2015. [Google Scholar]
- K Pal, NS Das, S Bhattacharya. Bacteriological profile of Indian currency circulating in a tertiary care hospital in rural Bengal. JRRMS 2013. [Google Scholar]
- Z Afroz. Bacteriological Profile and Antimicrobial Susceptibility Pattern of Indian Currency Circulating in a Tertiary Care Hospital of South India. Int J Curr Microbiol App Sci 2018. [Google Scholar]
- B Shrestha, BM Pokhrek, T Mohapatra. Phenotypic characterization of nososcomial isolates of Staphylococcus aureus with reference to MRSA. J Infect Dev Ctries 2009. [Google Scholar]
- S Ansari, H P Nepal, R Gautam, N Rayamajhi, S Shrestha, G Upadhyay. Threat of drug resistant Staphylococcus aureus to helth in Nepal. BMC Infect Dis 2014. [Google Scholar] [Crossref]
- NA Kassam, JD Damian, K Debora, N Balthazar, SK Gibson. Spectrum and antibiogram of bacteria isolated from patients presenting with infected wounds in a tertiary hospital, northern Tanzania. BMC Res Notes 2017. [Google Scholar] [Crossref]
- B Amabegaum, F Farooqui, P Ravichandra, Y K Sayeda, I Ifran. Aerobic bacteriological progile with antibiogram of pus isolates. Indian J Microbiol Res 2016. [Google Scholar]
- A Mohammed, M E Seid, T Gebrecherkos, M Tiruneh, F Moges. Bacterial Isolates and Their Antimicrobial Susceptibility Patterns of Wound Infections among Inpatients and Outpatients Attending the University of Gondar Referral Hospital, Northwest Ethiopia. Int J Microbiol 2017. [Google Scholar] [Crossref]
- A Belbase, DP Narayan, N Krishus, N Bihusan, B Rikesh, B Reena. Antibiotic resistance and biofilm production among the strains of Staphylococcus aureus isolated from pus /wound swab samples in a tertiary care hospital in Nepal. Ann Clin Microbiol Antimicrob 2017. [Google Scholar] [Crossref]
